JCM number: 1093 <-- T. Mitsuoka S8-9 <-- DSM 20196 <-- ATCC 10881 (Leuconostoc mesenteroides) <-- C. S. McCleskey 548-D.
Source: Sugar cane [018,5135].
Biochemistry/Physiology: [3808].
Cell wall: Lys-Ala [021,3808].
G+C (mol%): 45 (Tm) [020], 46.6 (HPLC) [2433].
DNA-DNA relatedness: [3808,5135].
Phylogeny: 16S rRNA (M23036), 16S rRNA gene (AB023241, AB596944, LC063164), [2983,3808,5135].
Other taxonomic data: Whole-cell protein profile [5135].
Genome sequence: JQAY00000000.
More information: Cellular polyamines [6386].
Genomic DNA is available from RIKEN BRC-DNA Bank: JGD19708.

open link in new window
open link in new window










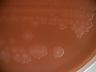
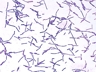


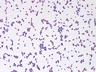




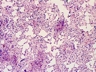










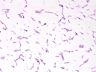





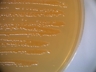


































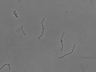


















































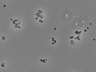
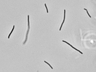
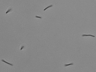
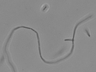












 Instructions for an order
Instructions for an order