JCM number: 1363 <-- K. Suzuki CNF 035 <-- AJ 1420 <-- ATCC 6946 <-- NCTC 4215 <-- H. L. Jensen.
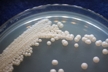
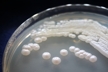
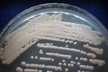
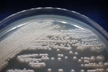
Source: Soil [274].
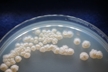
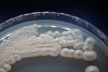
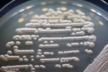
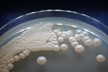
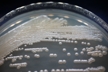
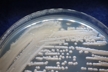
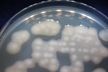
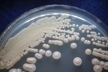
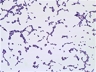
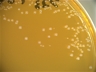
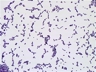
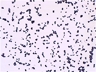
Morphology: [252].
Biochemistry/Physiology: [255,381].
Cell wall: LL-A2pm, Gly [377,378]; LL-A2pm-Gly [251,375]; N-acetyl Mur [276]; glycerol teichoic acid (Glc) [2883].
Fatty acid: [643,1015,1074,1090], contains 2OH acids [238,243,1012,1013,1014,1016,1017].
Quinone: MK-8(H4) [263,376,1074,1090].
Polar lipid: [264,1074,1090].
G+C (mol%): 74.0 [382], 71.7 (Tm) [125], 74.4 (Tm) [380], 73.0 (Tm) [381], 72.9 (genome sequence) [12227].
DNA-DNA relatedness: [259,381].
Phylogeny: 16S rRNA catalog [273], 16S rRNA (M37693), 16S rRNA gene (AF005009, X53213, Z78212) [2561,4327,5213], 5S rRNA (M16174) [2499], RNase P RNA gene (AF110040) [4865].
Other taxonomic data: Protein profile [223]; 16S-23S rRNA ITS (AF017494) [4385]; Rapid identification primers [4387]; Polyamine [4623].
Taxonomy: [250,256,274,388,1090,2561].
Genome sequence: BJMC00000000, FOUK00000000.
More information: Oxidizes cholesterol [383,384,385,386,387]; Carbohydrate catabolism [183].

open link in new window









































































































































































































































































 Instructions for an order
Instructions for an order