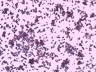
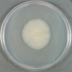
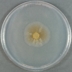
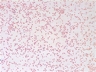
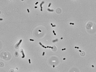
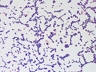
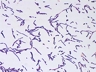
JCM number: 2010 <-- ATCC 33198 <-- VPI 1754.
Source: Hog small intestine.
Biochemistry/Physiology: [5147].
Enzyme electrophoretic profile: [3068].
G+C (mol%): 37.3 (Tm) [3068].
DNA-DNA relatedness: [1672,3068].
Phylogeny: 16S rRNA [2983], 16S rRNA gene (LC800112).
DNA typing: Ribotype [4925].
More information: Fermentation of glucose and ribose [4601]; Citrate lyase [4601]; Lactic acid production [5147]; 16S rRNA gene partial sequence of this strain has a high similarity (99.7%) with L. johnsonii.
Genomic DNA is available from RIKEN BRC-DNA Bank: JGD07980.
open link in new window

open link in new window




























 Instructions for an order
Instructions for an order