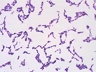
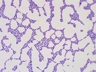
JCM number: 1217 <-- ATCC 15707 <-- G. Reuter E194b (Variant a).
Source: Intestine of adult [110,3126,6034].
Biochemistry/Physiology: [3093,3126,4424,5742,6034].
Cell wall: Orn-Ser-Ala-Thr-Ala [098,2925,3126].
Enzyme electrophoretic profile: [3126,4425].
G+C (mol%): 61.4 (Tm) [181], 61.3 (HPLC) [5742].
DNA-DNA relatedness: [023,053,3126,5742,6034].
Phylogeny: 16S rRNA (M58739), 16S rRNA gene (AB437359, GU361823, LC071818).
DNA typing: RFLP pattern [4423], rRNA gene restriction pattern [4597], ribotyping [6034], RAPD-PCR [6034].
Other taxonomic data: Protein profile [1865]; MALDI-TOF MS [9230].
Taxonomy: [3126].
Genome sequence: AP010888 [8490], FNRW00000000, JGYZ00000000, LR134369.
More information: Species-specific primer [4521]; endo-α-N-acetylgalactosaminidase [6623].
Genomic DNA is available from RIKEN BRC-DNA Bank: JGD07422.

open link in new window
open link in new window






























































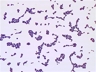

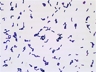

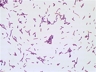



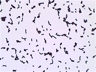


 Instructions for an order
Instructions for an order