Bacillus pinisoli
JCM number: 35212
<-- M.-G. Jiang and Y. Huang; Guangxi Univ. for Natl., China; GXH0341.
Distribution to users in Japan is controlled by the Plant Protection Law.
Source: Soil of a decayed pine tree in Weizhou Island, Beihai, Guangxi, China [14087].
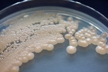
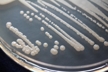
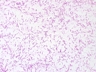
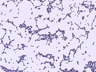
Morphology: [14087].
Biochemistry/Physiology: [14087].
Cell wall: meso-A2pm [14087].
Fatty acid: [14087].
Quinone: MK-7 [14087].
Polar lipid: DPG, PE, PG, PC, PLs [14087].
G+C (mol%): 37.0 (genome sequence) [14087].
Phylogeny: 16S rRNA gene (OL336472) [14087].
Other taxonomic data: Average nucleotide identity, genome-to-genome distances [14087].
Genome sequence: JAJQKI000000000 [14087].
|
| |

open link in new window
|


















 Instructions for an order
Instructions for an order